DeepChem
2 minute read
The DeepChem library provides open source tools that democratize the use of deep-learning in drug discovery, materials science, chemistry, and biology. This W&B integration adds simple and easy-to-use experiment tracking and model checkpointing while training models using DeepChem.
DeepChem logging in 3 lines of code
logger = WandbLogger(…)
model = TorchModel(…, wandb_logger=logger)
model.fit(…)
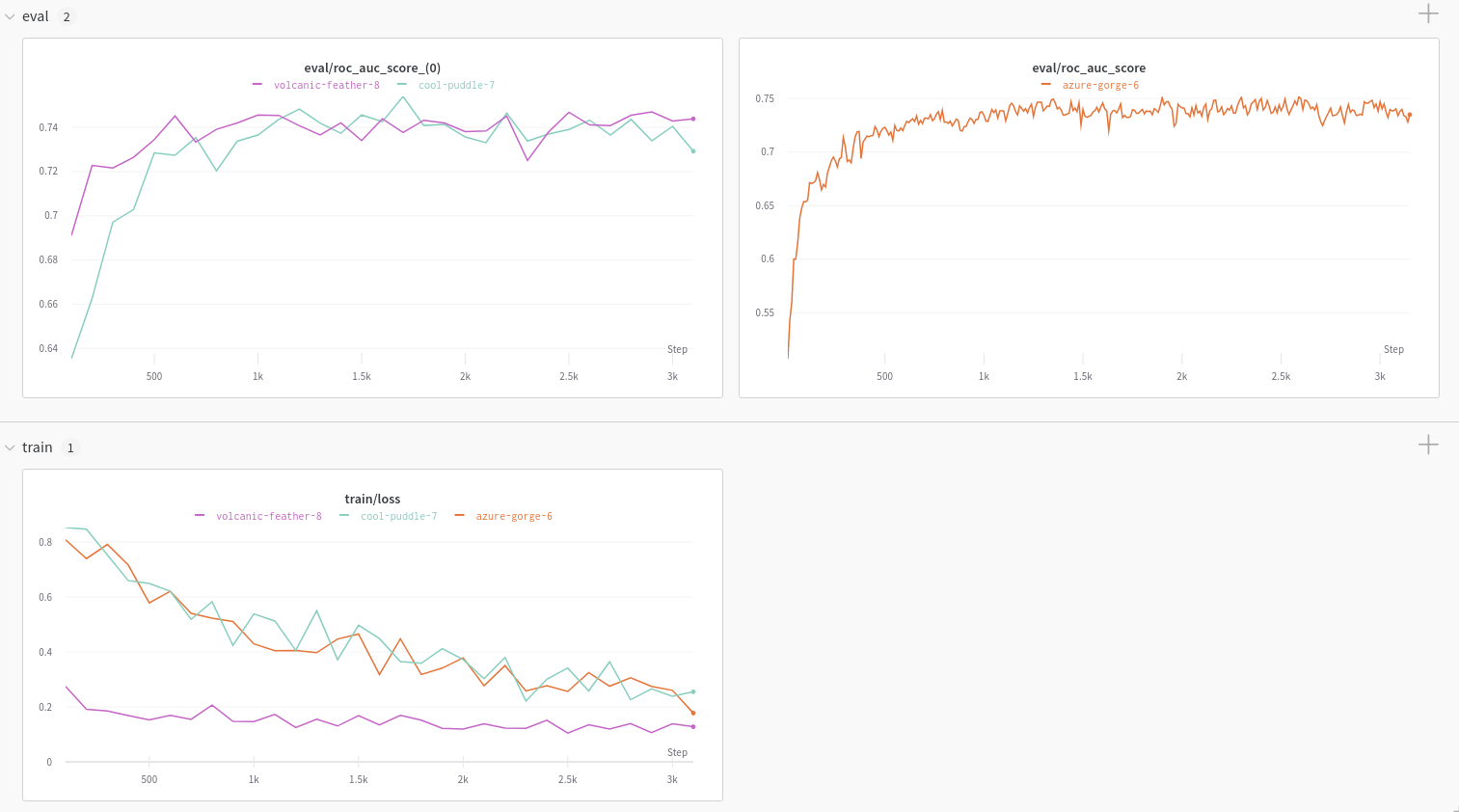
Report and Google Colab
Explore the Using W&B with DeepChem: Molecular Graph Convolutional Networks article for an example charts generated using the W&B DeepChem integration.
If you’d rather dive straight into working code, check out this Google Colab.
Track experiments
Setup Weights & Biases for DeepChem models of type KerasModel or TorchModel.
-
Install the
wandblibrary and log in``` pip install wandb wandb login ``````python !pip install wandb import wandb wandb.login() ``` -
Initialize and configure WandbLogger
from deepchem.models import WandbLogger logger = WandbLogger(entity="my_entity", project="my_project") -
Log your training and evaluation data to W&B
Training loss and evaluation metrics can be automatically logged to Weights & Biases. Optional evaluation can be enabled using the DeepChem ValidationCallback, the
WandbLoggerwill detect ValidationCallback callback and log the metrics generated.```python from deepchem.models import TorchModel, ValidationCallback vc = ValidationCallback(…) # optional model = TorchModel(…, wandb_logger=logger) model.fit(…, callbacks=[vc]) logger.finish() ``````python from deepchem.models import KerasModel, ValidationCallback vc = ValidationCallback(…) # optional model = KerasModel(…, wandb_logger=logger) model.fit(…, callbacks=[vc]) logger.finish() ```
Feedback
Was this page helpful?
Glad to hear it! Please tell us how we can improve.
Sorry to hear that. Please tell us how we can improve.